Applications & Publications
Technical Notes
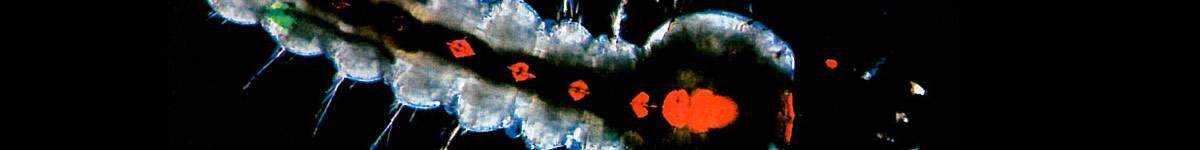
Sorting Embryos with Sex-Specific GFP-Expression (QTN-006)
Sex-Specific GFP-Expression in Embryos and Sorting using the COPAS Flow Cytometer (QTN-006)
Rapid Drosophila Embryo Sorting
Automated High Speed Analysis and Sorting of Drosophila Embryos (ANS-01)
Rare Drosophila Embryo Sorting
Isolation of Rare Flourescent Expressing Embryos from a Large Population (ANS-02)
Sorting Drosophila Imaginal Discs (QTN-005)
Publications
A Fluorescent Sex-Sorting Technique for Insects with the Demonstration in Drosophila melanogaster
Liu et al. February 15, 2024 GEN Biotechnol. February 2024; 3(1): 35–44. Published online 2024 Feb 15. doi: 10.1089/genbio.2023.0041
View AbstractA Fluorescent Sex-Sorting Technique for Insects with the Demonstration in Drosophila melanogaster
Recent advances in insect genetic engineering offer alternative genetic biocontrol solutions to control populations of pests and disease vectors. While success has been achieved, sex-sorting remains problematic for scaling many genetic biocontrol interventions. Here, we describe the development of a genetically stable sex-sorting technique for female and male selection with a proof of concept in Drosophila melanogaster termed SEPARATOR (Sexing Element Produced by Alternative RNA-splicing of A Transgenic Observable Reporter). This elegant approach utilizes dominantly expressed fluorescent proteins and differentially spliced introns to ensure sex-specific expression. The system has the potential for adaptability to various insect species and application for high-throughput insect sex-sorting.
Chromatin accessibility in the Drosophila embryo is determined by transcription factor pioneering and enhancer activation
Brennan et al. October 09, 2023 Dev Cell. Author manuscript; available in PMC 2023 Oct 23.Published in final edited form as: Dev Cell. 2023 Oct 9; 58(19): 1898–1916.e9. Published online 2023 Aug 8. doi: 10.1016/j.devcel.2023.07.007
View AbstractChromatin accessibility in the Drosophila embryo is determined by transcription factor pioneering and enhancer activation
Chromatin accessibility is integral to the process by which transcription factors (TFs) read out cis-regulatory DNA sequences, but it is difficult to differentiate between TFs that drive accessibility and those that do not. Deep learning models that learn complex sequence rules provide an unprecedented opportunity to dissect this problem. Using zygotic genome activation in Drosophila as a model, we analyzed high-resolution TF binding and chromatin accessibility data with interpretable deep learning and performed genetic validation experiments. We identify a hierarchical relationship between the pioneer TF Zelda and the TFs involved in axis patterning. Zelda consistently pioneers chromatin accessibility proportional to motif affinity, whereas patterning TFs augment chromatin accessibility in sequence contexts where they mediate enhancer activation. We conclude that chromatin accessibility occurs in two tiers: one through pioneering, which makes enhancers accessible but not necessarily active, and the second when the correct combination of TFs leads to enhancer activation.
A fluorescent sex-sorting technique for insects with the demonstration in Drosophila melanogaster
Liu et al. August 14, 2023 Version 1. bioRxiv. Preprint. 2023 Aug 14. doi: 10.1101/2023.08.11.553026Published in: GEN Biotechnol. February 2024; 3(1): 35–44.
View AbstractA fluorescent sex-sorting technique for insects with the demonstration in Drosophila melanogaster
Recent advances in insect genetic engineering offer alternative genetic biocontrol solutions to control populations of pests and disease vectors. While success has been achieved, sex-sorting remains problematic for scaling many genetic biocontrol interventions. Here we describe the development of a sex-sorting technique for female and male selection with a proof-of-concept in D. melanogaster termed SEPARATOR (Sexing Element Produced by Alternative RNA-splicing of A Transgenic Observable Reporter). This approach utilizes dominant fluorescent proteins and differentially spliced introns to ensure sex-specific expression. The system has the potential for adaptability to various insect species and application for high-throughput insect sex-sorting.
Nuclear elongation during spermiogenesis depends on physical linkage of nuclear pore complexes to bundled microtubules by Drosophila Mst27D
Li et al. July 10, 2023 PLoS Genet. 2023 Jul; 19(7): e1010837. Published online 2023 Jul 10. doi: 10.1371/journal.pgen.1010837
View AbstractNuclear elongation during spermiogenesis depends on physical linkage of nuclear pore complexes to bundled microtubules by Drosophila Mst27D
Spermatozoa in animal species are usually highly elongated cells with a long motile tail attached to a head that contains the haploid genome in a compact and often elongated nucleus. In Drosophila melanogaster, the nucleus is compacted two hundred-fold in volume during spermiogenesis and re-modeled into a needle that is thirty-fold longer than its diameter. Nuclear elongation is preceded by a striking relocalization of nuclear pore complexes (NPCs). While NPCs are initially located throughout the nuclear envelope (NE) around the spherical nucleus of early round spermatids, they are later confined to one hemisphere. In the cytoplasm adjacent to this NPC-containing NE, the so-called dense complex with a strong bundle of microtubules is assembled. While this conspicuous proximity argued for functional significance of NPC-NE and microtubule bundle, experimental confirmation of their contributions to nuclear elongation has not yet been reported. Our functional characterization of the spermatid specific Mst27D protein now resolves this deficit. We demonstrate that Mst27D establishes physical linkage between NPC-NE and dense complex. The C-terminal region of Mst27D binds to the nuclear pore protein Nup358. The N-terminal CH domain of Mst27D, which is similar to that of EB1 family proteins, binds to microtubules. At high expression levels, Mst27D promotes bundling of microtubules in cultured cells. Microscopic analyses indicated co-localization of Mst27D with Nup358 and with the microtubule bundles of the dense complex. Time-lapse imaging revealed that nuclear elongation is accompanied by a progressive bundling of microtubules into a single elongated bundle. In Mst27D null mutants, this bundling process does not occur and nuclear elongation is abnormal. Thus, we propose that Mst27D permits normal nuclear elongation by promoting the attachment of the NPC-NE to the microtubules of the dense complex, as well as the progressive bundling of these microtubules.
A high throughput method for egg size measurement in Drosophila
Barghi et al. March 07, 2023 Drosophila. Sci Rep 13, 3791 (2023). https://doi.org/10.1038/s41598-023-30472-8
A high throughput method for egg size measurement in Drosophila
Mass Purification Protocol for Drosophila melanogaster Wing Imaginal Discs: An Alternative to Dissection to Obtain Large Numbers of Disc Cells
Hoareau et al. September 22, 2022 Biology (Basel) 2022 Oct; 11(10): 1384. Published online 2022 Sep 22. doi:Â 10.3390/biology11101384
View AbstractMass Purification Protocol for Drosophila melanogaster Wing Imaginal Discs: An Alternative to Dissection to Obtain Large Numbers of Disc Cells
Drosophila melanogaster imaginal discs are larval internal structures that become the external organs of the adult. They have been used to study numerous developmental processes for more than fifty years. Dissecting these imaginal discs for collection is challenging, as the size of third-instar larvae organs is typically less than 1 mm. Certain experimental applications of the organs require many cells, which requires researchers to spend several hours dissecting them. This paper proposes an alternative to dissection in the form of a mass enrichment protocol. The protocol enables the recovery of many wing imaginal discs by grinding large quantities of third-instar larvae and separating the organs using filtration and a density gradient. The wing imaginal discs collected with this protocol in less than three hours are as well preserved as those collected by dissection. The dissociation and filtration of the extract allow the isolation of a large amount of wing imaginal disc cells.
Ribbon boosts ribosomal protein gene expression to coordinate organ form and function
Loganathan et al. February 23, 2022 J Cell Biol. 2022 Apr 4; 221(4): e202110073. Published online 2022 Feb 23. doi: 10.1083/jcb.202110073
View AbstractRibbon boosts ribosomal protein gene expression to coordinate organ form and function
Cell growth is well defined for late (postembryonic) stages of development, but evidence for early (embryonic) cell growth during postmitotic morphogenesis is limited. Here, we report early cell growth as a key characteristic of tubulogenesis in the Drosophila embryonic salivary gland (SG) and trachea. A BTB/POZ domain nuclear factor, Ribbon (Rib), mediates this early cell growth. Rib binds the transcription start site of nearly every SG-expressed ribosomal protein gene (RPG) and is required for full expression of all RPGs tested. Rib binding to RPG promoters in vitro is weak and not sequence specific, suggesting that specificity is achieved through cofactor interactions. Accordingly, we demonstrate Rib's ability to physically interact with each of the three known regulators of RPG transcription. Surprisingly, Rib-dependent early cell growth in another tubular organ, the embryonic trachea, is not mediated by direct RPG transcription. These findings support a model of early cell growth customized by transcriptional regulatory networks to coordinate organ form and function.
Chromosome separation during Drosophila male meiosis I requires separase-mediated cleavage of the homolog conjunction protein UNO
Weber at al. October 01, 2020 PLoS Genet. 2020 Oct; 16(10): e1008928. Published online 2020 Oct 1. doi: 10.1371/journal.pgen.1008928
View AbstractChromosome separation during Drosophila male meiosis I requires separase-mediated cleavage of the homolog conjunction protein UNO
Defects in the Neuroendocrine Axis Contribute to Global Development Delay in a Drosophila Model of NGLY1 Deficiency
Rodriguez et al. May 07, 2018 https://doi.org/10.1534/g3.118.300578 Manuscript received January 8, 2018; accepted for publication April 17, 2018; published Early Online May 7, 2018.
Defects in the Neuroendocrine Axis Contribute to Global Development Delay in a Drosophila Model of NGLY1 Deficiency
Combining Time-of-Flight Secondary Ion Mass SpectrometryImaging Mass Spectrometry and CARS Microspectroscopy Reveals Lipid Patterns Reminiscent of Gene Expression Patterns in the Wing Imaginal Disc of Drosophila melanogaster
Marty et al. July 20, 2017 DOI: 10.1021/acs.analchem.7b00125, Received: January 11, 2017, Accepted: July 20, 2017, Published: July 20, 2017
Combining Time-of-Flight Secondary Ion Mass SpectrometryImaging Mass Spectrometry and CARS Microspectroscopy Reveals Lipid Patterns Reminiscent of Gene Expression Patterns in the Wing Imaginal Disc of Drosophila melanogaster
A large-scale, in vivo transcription factor screen defines bivalent chromatin as a key property of regulatory factors mediating Drosophila wing development.
Schertel C¹, Albarca M, Rockel-Bauer C¹, Kelley NW, Bischof J¹, Hens K, van Nimwegen E, Basler K¹, Deplancke B¹. April 25, 2015 Genome Res. 2015 Apr;25(4):514-23. doi: 10.1101/gr.181305.114. Epub 2015 Jan 7.
View AbstractA large-scale, in vivo transcription factor screen defines bivalent chromatin as a key property of regulatory factors mediating Drosophila wing development.
1Institute of Molecular Life Sciences, University of Zurich, 8057 Zurich, Switzerland
Large-scale imaginal disc sorting: a protocol for "omics"-approaches.
Marty F, Rockel-Bauer C, Simigdala N, Brunner E, Basler K. April 12, 2014 Methods. 2014 Apr 12. pii: S1046-2023(14)00149-2. doi: 10.1016/j.ymeth.2014.04.005. [Epub ahead of print]
View AbstractLarge-scale imaginal disc sorting: a protocol for "omics"-approaches.
Institute of Molecular Life Sciences, University of Zurich, Winterthurerstrasse 190, 8057 Zurich, CH.
Small molecule drug screening in Drosophila identifies the 5HT2A receptor as a feeding modulation target.
Gasque G, Conway S, Huang J, Rao Y, Vosshall LB. July 02, 2013 Sci Rep 2013;3:srep02120.doi:10.1038/srep02120
View AbstractSmall molecule drug screening in Drosophila identifies the 5HT2A receptor as a feeding modulation target.
The Rockefeller University, 1230 York Avenue, Box 63, New York, NY 10065, U.S.A.
Genome-Wide Analysis Reveals a Major Role in Cell Fate Maintenance and an Unexpected Role in Endoreduplication for the Drosophila FoxA Gene Fork Head
Rika Maruyama, Elizabeth Grevengoed, Peter Stempniewicz, Deborah J. Andrew June 16, 2011 PLoS ONE 6(6): e20901. doi:10.1371/journal.pone.0020901
View AbstractGenome-Wide Analysis Reveals a Major Role in Cell Fate Maintenance and an Unexpected Role in Endoreduplication for the Drosophila FoxA Gene Fork Head
Department of Cell Biology, The Johns Hopkins University, School of Medicine, Baltimore, Maryland, United States of America
The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity
Rebecca M. Fox, Caitlin D. Hanlon, and Deborah J. Andrew November 01, 2010 Published November 1, 2010 // JCB vol. 191 no. 3 479-492
View AbstractThe CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity
Department of Cell Biology, The Johns Hopkins University, School of Medicine, Baltimore, MD 21205
Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication.
Yi X, de Vries HI, Siudeja K, Rana A, Lemstra W, Brunsting JF, Kok RM, Smulders YM, Schaefer M, Dijk F, Shang Y, Eggen BJ, Kampinga HH, Sibon OC November 20, 2009
View AbstractStwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication.
Department of Radiation and Stress Cell Biology, Division of Cell Biology, University Medical Center Groningen, University of Groningen, 9713 AV Groningen, The Netherlands.
Abstract:
Hydroxyurea, a well-known DNA replication inhibitor, induces cell cycle arrest and intact checkpoint functions are required to survive DNA replication stress induced by this genotoxic agent. Perturbed DNA synthesis also results in elevated levels of DNA damage. It is unclear how organisms prevent accumulation of this type of DNA damage that coincides with hampered DNA synthesis. Here, we report the identification of stonewall (stwl) as a novel hydroxyurea-hypersensitive mutant. We demonstrate that Stwl is required to prevent accumulation of DNA damage induced by hydroxyurea; yet, Stwl is not involved in S/M checkpoint regulation. We show that Stwl is a heterochromatin-associated protein with transcription-repressing capacities. In stwl mutants, levels of trimethylated H3K27 and H3K9 (two hallmarks of silent chromatin) are decreased. Our data provide evidence for a Stwl-dependent epigenetic mechanism that is involved in the maintenance of the normal balance between euchromatin and heterochromatin and that is required to prevent accumulation of DNA damage in the presence of DNA replication stress.
In vivo proteomics in Drosophila melanogaster by tandem affinity purification of protein complexes and analysis by Protein Center software
ABRF 2008, Feb 9-12, Salt Lake City, Utah
J.S. Rees1 , M. BBern2 , D. St Johnston1, K. Lilley1
February 09, 2008
In vivo proteomics in Drosophila melanogaster by tandem affinity purification of protein complexes and analysis by Protein Center software
1University of Cambridge, Cambride, UK; 2Proxeon, Odense, Denmark
Genome-wide mapping and characterisation of protein expression and interaction in Drosophila melanogaster, using a hybrid piggyBac/P-element YFP gene trap system with tandem affinity tags
48th Annual Drosophila Research Conference, March 7 – 11, 2007
E. Ryder1, H. Spriggs1, G. Johnson1, E. Drummond1, J. Drummond1, J. Webster1,
J. Roote1, N. Lowe2, K. Lilley3, S. Hester3, J. Howard3, J. Rees3, S. Russell1, 3, D. St. Johnston2.
March 07, 2007
Genome-wide mapping and characterisation of protein expression and interaction in Drosophila melanogaster, using a hybrid piggyBac/P-element YFP gene trap system with tandem affinity tags
1. Department of Genetics, University of Cambridge, Downing Street, Cambridge, CB2 3EH. UK. 2. The Wellcome Trust/Cancer Research UK Gurdon Institute, University of Cambridge, Tennis Court Road, Cambridge CB2. 1QN. 3. Cambridge Systems Biology Centre, Tennis Court Road, Cambridge, CB2 1QR
Exploring Strategies for Protein Trapping in Drosophila
Ana T. Quiñones-Coello*,1, Lisa N. Petrella*,1, Kathleen Ayers*,1, Anthony Melillo*, Stacy Mazzalupo*,2, Andrew M. Hudson*, Shu Wang*,3, Claudia Castiblanco*, Michael Buszczak†,‡, Roger A. Hoskins§ and Lynn Cooley*,†,**,4
March 01, 2007
Genetics, Vol. 175, 1089-1104, March 2007, Copyright © 2007
doi:10.1534/genetics.106.065995
Exploring Strategies for Protein Trapping in Drosophila
* Department of Genetics, ** Department of Cell Biology and †Department of Molecular, Cellular and Developmental Biology, Yale University School of Medicine, New Haven, Connecticut 06520, ‡Department of Embryology, Carnegie Institution of Washington, Baltimore, Maryland 21218 and §Department of Genome Biology, Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720
The Carnegie Protein Trap Library: A Versatile Tool for Drosophila Developmental Studies
Michael Buszczak*, Shelley Paterno*, Daniel Lighthouse*, Julia Bachman*, Jamie Planck*, Stephenie Owen*, Andrew D. Skora*, Todd G. Nystul*, Benjamin Ohlstein*, Anna Allen*, James E. Wilhelm*, Terence D. Murphy*, Robert W. Levis*, Erika Matunis†, Nahathai Srivali*, Roger A. Hoskins‡ and Allan C. Spradling*,1
March 01, 2007
Genetics, Vol. 175, 1505-1531, March 2007, Copyright © 2007
doi:10.1534/genetics.106.065961
The Carnegie Protein Trap Library: A Versatile Tool for Drosophila Developmental Studies
*Howard Hughes Medical Institute Research Laboratories, Department of Embryology, Carnegie Institution of Washington, Baltimore, Maryland 21218, †Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205 and ‡Department of Genome Biology, Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720
An efficient promoter trap for detection of patterned gene expression and subsequent functional analysis in Drosophila.
Larsen C, Franch-Marro X, Hartenstein V, Alexandre C, Vincent JP. November 21, 2006 Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17813-7. Epub 2006 Nov 8.
An efficient promoter trap for detection of patterned gene expression and subsequent functional analysis in Drosophila.
Microarray-based screens to identify genes specific to the fusion-competent myoblasts. (874A) (Poster)
46th Annual Drosophila Research Conference, March 30 – April 3, 2005
Shruti Haralalka, Susan Abmayr.
March 30, 2005
Microarray-based screens to identify genes specific to the fusion-competent myoblasts. (874A) (Poster)
Stowers Institute for Medical Research, Kansas City, MO 64110.
Gene expression profiling during gliogenesis in Drosophila.
46th Annual Drosophila Research Conference, March 30 – April 3, 2005
Becker A.
March 30, 2005
Gene expression profiling during gliogenesis in Drosophila.
Institute of Genetics, University of Mainz, Germany
High throughput collection of Drosophila embryos for homozygous lethal mutants based on deformed driven YFP expression
46th Annual Drosophila Research Conference, March 30 – April 3, 2005
Bo Wang1, Julia Thompson1, Greg Beitel2 and Rock Pulak1
March 30, 2005
High throughput collection of Drosophila embryos for homozygous lethal mutants based on deformed driven YFP expression
1) Union Biometrica Inc., 84 October Hill Road, Holliston, MA 01746 USA; 2) Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, IL 60208.
Sex-specific GFP-expression in Drosophila embryos and sorting by COPAS flow cytometry technique (#615C).
45th Annual Drosophila Research Conference, March 24-28, 2004
Julia Thompson1, Patricia Graham2, Paul Schedl2 and Rock Pulak1
March 24, 2004
Sex-specific GFP-expression in Drosophila embryos and sorting by COPAS flow cytometry technique (#615C).
1) Union Biometrica Inc (Holliston, MA USA), 2) Department of Molecular Biology, Princeton University (Princeton, NJ USA).
Drosophila Fluorescent-Imaginal Disc Analysis and Sorting by COPAS Flow Cytometry Technique (412A).
45th Annual Drosophila Research Conference, March 24-28, 2004
Daniela Panáková1, Lydia Michaut2, Rico Bongaarts3, Bo Wang4, Julia Thompson4, Rock Pulak4
March 24, 2004
Drosophila Fluorescent-Imaginal Disc Analysis and Sorting by COPAS Flow Cytometry Technique (412A).
1) Max Planck Institute, CBG, Susan Eaton Lab (Dresden, Germany), 2) Biozentrum, Walter Gehring lab (Basel, Switzerland), 3) Union Biometrica (Geel, Belgium), 4) Union Biometrica Inc. (Holliston, MA, USA).
Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster.
Reed J. Kelso, Michael Buszczak1, Ana T. QuinÄones2, Claudia Castiblanco2, Stacy Mazzalupo2 and Lynn Cooley2,3,* January 01, 2004 Nucleic Acids Research (2004) Vol. 32, Database issue Department of Cellular and Molecular Pharmacology, Howard Hughes Medical Institute, University of California, San Francisco, CA 94143, USA 1) Department of Embryology, Carnegie Institution of Washington, Baltimore, MD 21210, USA 2) Department of Genetics, and 3) Department of Cell Biology, Yale University School of Medicine, New Haven, CT 06520-8005, USA
View AbstractFlytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster.
Flytrap is a web-enabled relational database of transposable element insertions in Drosophila melanogaster . A green fluorescent protein (GFP) artificial exon carried by a transposable P -element is mobilized and inserted into a host gene intron creating a GFP fusion protein. The sequence of the tagged gene is determined by sequencing inverse-PCR products derived from genomic DNA. Flytrap contains two principle data types: micrographs of protein localization and a cellular component ontology, based on rules derived from the Gene Ontology consortium ( http://www.geneontology.org ), describing protein localization. Flytrap also has links to gene information contained in Flybase ( http:// flybase.bio.indiana.edu ). The system is designed to accept submissions of micrographs and descriptions from any type of tissue (e.g. wing imaginal disk, ovary) and at any stage of development. Insertion lines can be searched using a number of queries, including Berkeley Drosophila Genome Project (BDGP) numbers and protein localization. In addition, Flytrap provides online order forms linked to each insertion line so that users may request any line generated from this project. Flytrap may be accessed from the homepage at http://flytrap.med. yale.edu .
A report on the 62nd Annual Meeting for the Society for Developmental Biology (SDB), Boston, USA, 30 July to 3 August 2003, "Developments in developmental genomics."
Holly A Field1 and Kevin P White2 July 30, 2003 Genome Biology 2003, 4:345 1) Department of Biochemistry and Biophysics, University of California San Francisco, 513 Parnassus Avenue, San Francisco, CA 94143, USA; 2) Department of Genetics, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520, USA
View AbstractA report on the 62nd Annual Meeting for the Society for Developmental Biology (SDB), Boston, USA, 30 July to 3 August 2003, "Developments in developmental genomics."
Developmental biologists from around the world converged on Boston for the SDB annual meeting to discuss topics that ranged from embryonic development in invertebrates to mammalian stem cells. Genomic approaches are driving many new discoveries in developmental biology, as was reflected in several plenary-session talks and a special workshop that featured genomic and proteomic applications. A few of the highlights are described here.
Instrumentation for analysis and sorting of fluorescence patterns in transgenic Drosophila embryos, #1001B.
44th Annual Drosophila Research Conference, March 5-9, 2003
R. Pulak, B. Moellers, J. Thompson, B. Wang
March 05, 2003
Instrumentation for analysis and sorting of fluorescence patterns in transgenic Drosophila embryos, #1001B.
Life Sciences, Union Biometrica, Inc. (Holliston, MA.)
Binding site for p120/-catenin is not required for Drosophila E-cadherin function in vivo.
Anne Pacquelet, Li Lin and Pernille Rørth January 27, 2003 The Journal of Cell Biology (2003) Vol. 160, # 3, pp. 313-319 European Molecular Biology Laboratory, 69117 Heidelberg, Germany
View AbstractBinding site for p120/-catenin is not required for Drosophila E-cadherin function in vivo.
Homophilic cell adhesion mediated by classical cadherins is important for many developmental processes. Proteins that interact with the cytoplasmic domain of cadherin, in particular the catenins, are thought to regulate the strength and possibly the dynamics of adhesion. β-catenin links cadherin to the actin cytoskeleton via α-catenin. The role of p120/δ-catenin proteins in regulating cadherin function is less clear. Both β-catenin and p120/δ-catenin are conserved in Drosophila. Here, we address the importance of cadherin–catenin interactions in vivo, using mutant variants of Drosophila epithelial cadherin (DE-cadherin) that are selectively defective in p120ctn (DE-cadherin-AAA) or β-catenin–armadillo (DE-cadherin-Δβ) interactions. We have analyzed the ability of these proteins to substitute for endogenous DE-cadherin activity in multiple cadherin-dependent processes during Drosophila development and oogenesis; epithelial integrity, follicle cell sorting, oocyte positioning, as well as the dynamic adhesion required for border cell migration. As expected, DE-cadherin-Δβ did not substitute for DE-cadherin in these processes, although it retained some residual activity. Surprisingly, DE-cadherin-AAA was able to substitute for the wild-type protein in all contexts with no detectable perturbations. Thus, interaction with p120/δ-catenin does not appear to be required for DE-cadherin function in vivo.
High Throughput Protein Trapping in Drosophila, #982A.
43rd Annual Drosophila Research Conference, April 10-14, 2002
M.H. Buszczak1, X. Morin2, A.T. Quinones 3, W. Chia 2, L. Cooley 3,4
April 10, 2002
High Throughput Protein Trapping in Drosophila, #982A.
Dept MCDB, Yale Univ, New Haven, CT; 2) Institute of Molecular and Cell Biology, King's College London, Guy's Hospital, London, UK; 3) Dept Genetics, Yale Univ, New Haven, CT; 4) Dept Cell Biology, Yale Univ, New Haven, CT.
Using Protein Traps to Study Genes Expressed during Oogenesis, #608B.
43rd Annual Drosophila Research Conference, April 10-14, 2002
A.T. Quiñones1, M. Buszczak1, X. Morin2, W. Chia2, L. Cooley1
April 10, 2002
Using Protein Traps to Study Genes Expressed during Oogenesis, #608B.
1) Genetics, Yale University, New Haven, CT; 2) MRC Centre for Developmental Neurobiology, King's College London, New Hunts House, Guy's Hospital, London, UK.
A rapid and efficient approach to vital enhancer trap screening in Drosophila embryos, #143.
43rd Annual Drosophila Research Conference, April 10-14, 2002
S.S. Gisselbrecht1, J. Bayes1, J. Etchin1, B. Dell'Orfano2, A. Ferrante2, A.M. Michelson1
April 10, 2002
A rapid and efficient approach to vital enhancer trap screening in Drosophila embryos, #143.
Howard Hughes Medical Institute, Brigham and Women's Hospital and Harvard Medical School, Boston, MA; 2) Union Biometrica, Inc., Holliston, MA.
Developing a Drosophila high throughput screen technology, #949A.
42nd Annual Drosophila Research Conference, March 21-25, 2001
Li, H.H., Wang, Q., Shi, X. and Zusman, S.
March 21, 2001
Developing a Drosophila high throughput screen technology, #949A.
S. Dept. of Functional Genomics, Novartis Biomedical Research Institute (Summit NJ)
Fully Automated Instrumentation for Gene Function Analysis in D. melanogaster, #956B (p. a330).
42nd Annual Drosophila Research Conference, March 21-25, 2001
Kalutkiewicz, P., Holcombe, B., Clover, R. and Chouinard, S.*
March 21, 2001
Fully Automated Instrumentation for Gene Function Analysis in D. melanogaster, #956B (p. a330).
Union Biometrica, Inc. (Holliston, MA), *Cambria Biosciences, LLC (Bedford, MA)